Our Genomic Analysis Approach
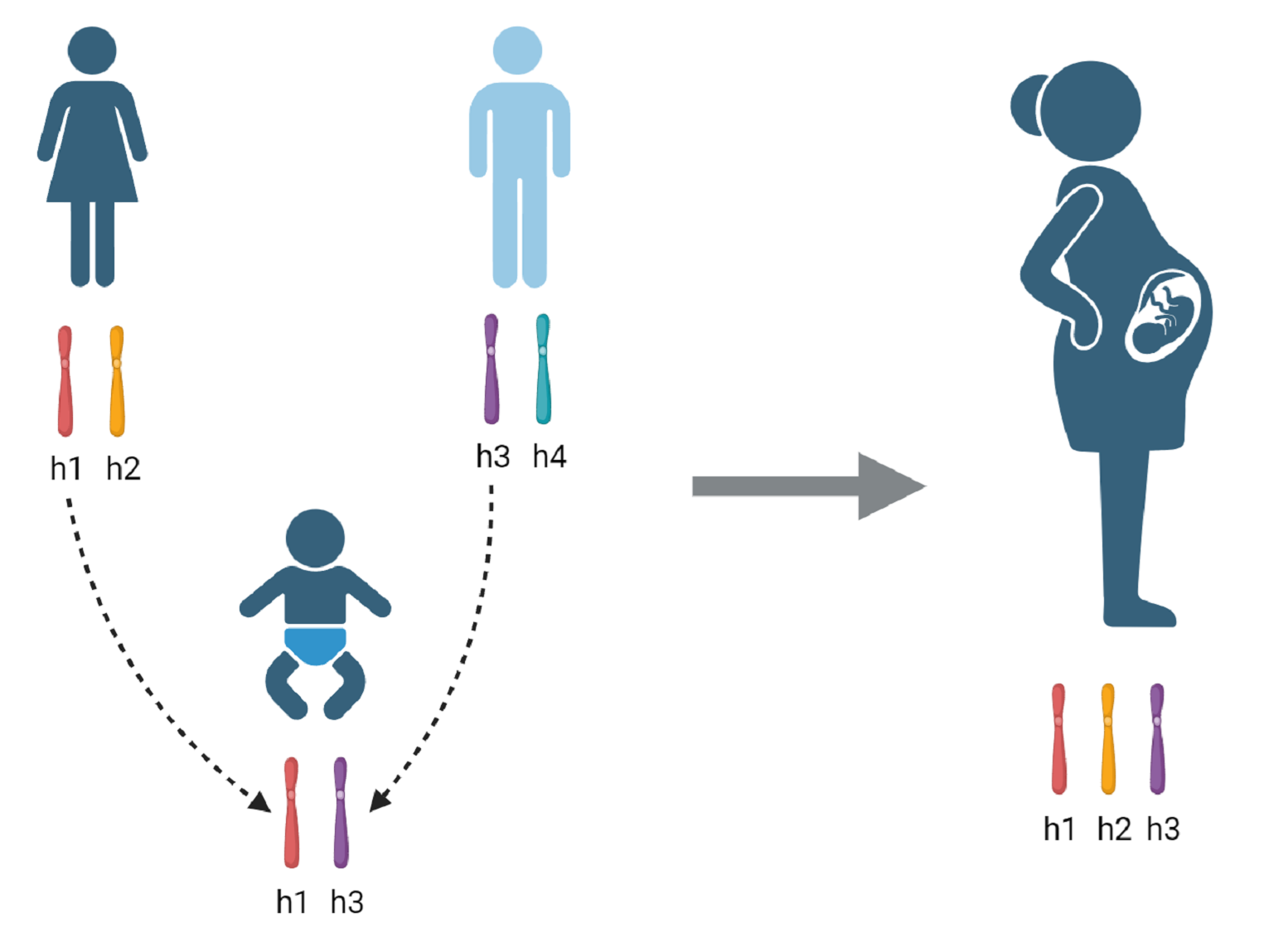
Our lab uses an integrated strategy to disentangle how pregnancy outcomes arise from maternal, paternal, and fetal genetic effects. By leveraging genomic data in mother–child pairs (and, where possible, paternal genotypes), we can systematically separate the influence of the maternal genome from direct fetal effects. This framework enables us to identify genetic variants that shape gestational duration, birth weight, and other pregnancy traits more precisely, guiding both mechanistic studies and potential clinical applications.
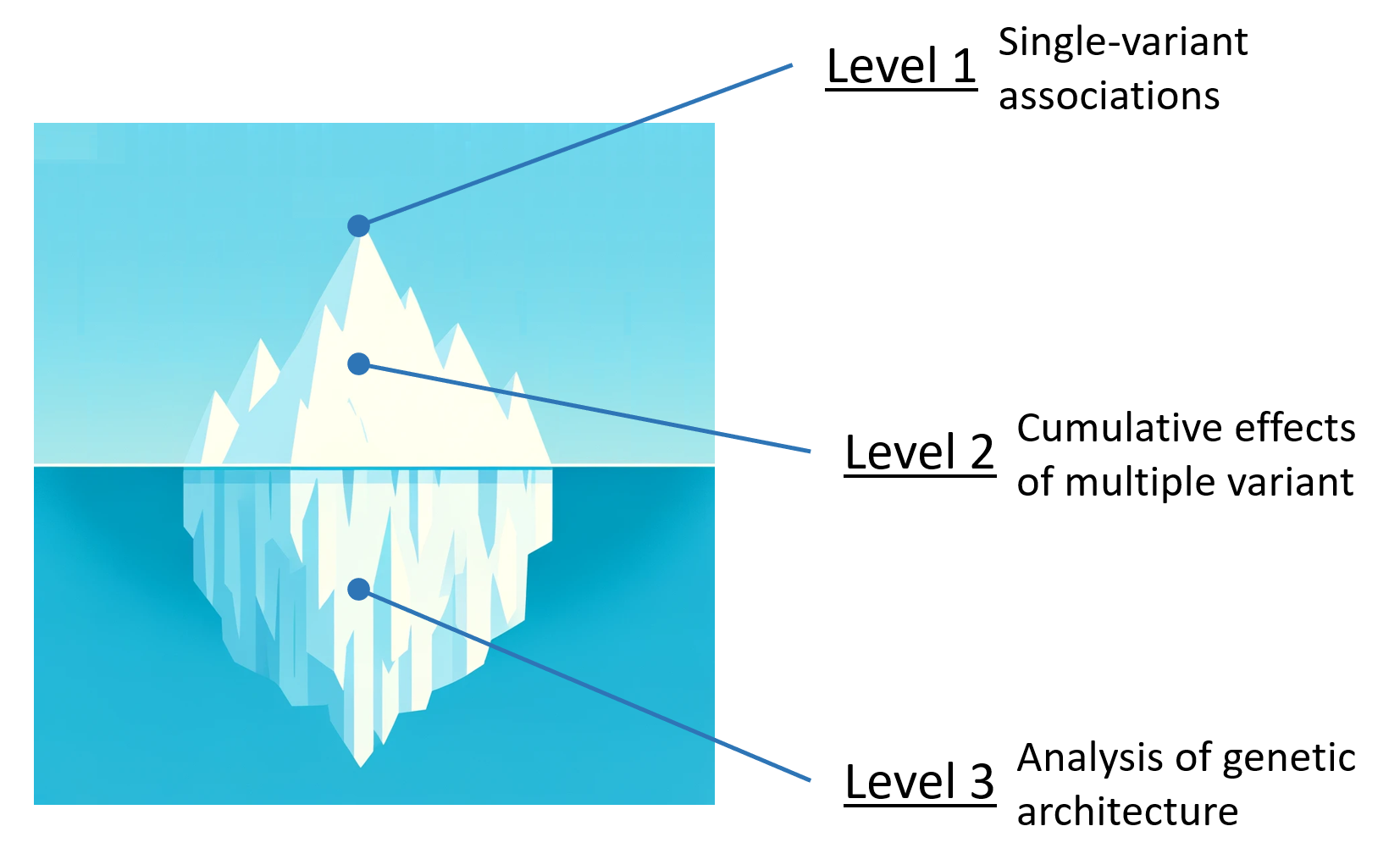
We further examine the genetics of pregnancy outcomes at three complementary levels. Level 1 captures single-variant associations, highlighting individual loci that may affect birth outcomes. Level 2 focuses on the cumulative effects of multiple variants, providing a polygenic view of risk and resilience. Finally, Level 3 digs deeper into the overall genetic architecture—investigating how variants interact with each other and with environmental exposures to influence pregnancy phenotypes across diverse populations.
By integrating these levels of analysis within our mother–child framework, we gain a more holistic view of how genetic and environmental factors combine to drive pregnancy outcomes. This approach not only pinpoints actionable gene targets for further functional validation and clinical translation but also illuminates the broader mechanistic pathways linking pregnancy biology to long-term health.